|
The iSect
Toolbox:
The iSect Tools is a collection of
the integrated tools in the Genome Express Browser System (GEBrowser) to
allow users to manipulate
the genomics
database, to retrieve genomic data and sequences, to designed suitable PCR
primers and to carry out gene expression
data mining.
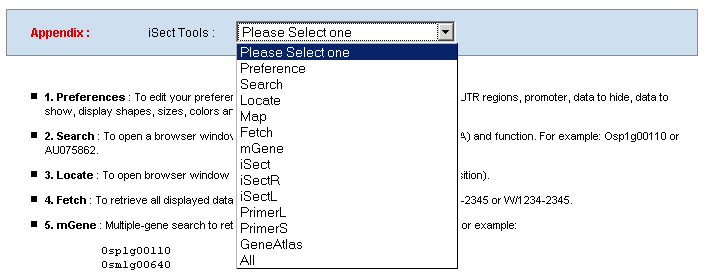
1. Preferences: To edit your preference to set your own graphic display and database mining parameters, like image width, zoom factor, UTR regions, promoter, data to hide, data to show, display shapes, sizes, colors and track spans and etc. .  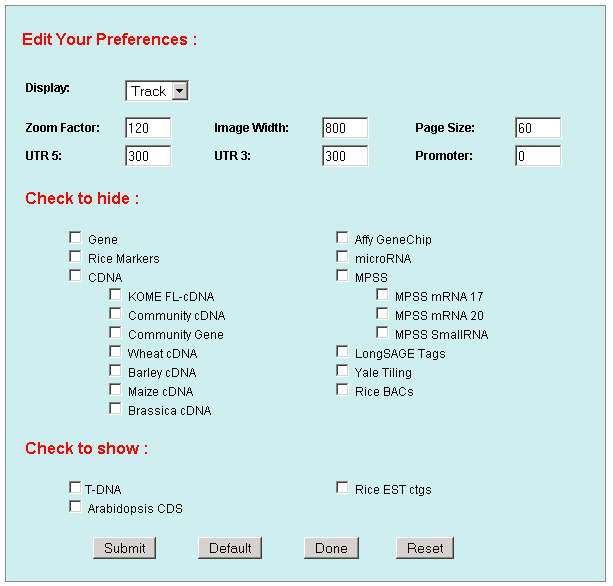  a. Display: including Track (multiple-track) display and Tiling (Duo-track) display. The Tiling display will ignore any histogram or TOME data. b. Zoom factor: the base number of a pixel distance in the displayed image map. c. Image width: the width of the working area in the displayed image. d. Page size: the number of display entries in the function search or the gAtlas display. e. UTR5’, UTR3’ and Promoter: the average lengths of UTR5’, UTR3’ and Promoters of genes in the whole genome. 2. Search: To open a browser window by querying a gene name, a BAC name, or a data name ( like cDNA, T-DNA or other). Or search the gene annotation or functions. For example:
a. To search a gene "At1g23450" in Arabidopsis database or "Osp1g00110" in RiceGE, you can select the drowpdown
menu of "Type:" as "Gene" and input "At1g23450" or "Osp1g00110" in the "Qurey" field.
b. To query a T-DNA line or cDNA clone, like SALK_000002 or AU075862, you can set the "Type" as "Data", input
SALK_000002 or AU075862 in the "Qurey" field.
c. Set the "Type" as "Function" to query genes which annotations contain your input keywords, like auxin.  3. Locate: To open browser window by positioning its location (chromosome and position).  4. Fetch: To retrieve all displayed data entries from a chromosome region. For example: 1234-2345 or W/1234-2345. 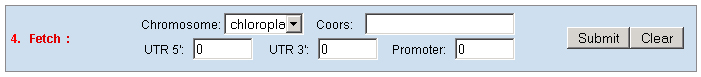 5. mText: Multiple-gene or multiple T-DNA/cDNA search to allow retrieving data or genes related a list of genes/T-DNAs/cDNAs. For example: Osp1g00110 Osm1g00640 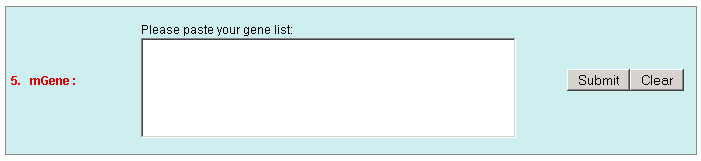 6. iSectR: iSect (R) tool to retrieve a fragments of sequences (sequences in a specific chromosome region) from the pseudo-molecules For example: chr01 [chromosome] and 1234-2345 or 1234-2345,3456-4567 [Range or Coordinates]. 7. iSectL: iSect (L) tool to retrieve a fragments of sequences from the genome corresponding to the mapping coordinates of a list of
existed data names in the database or a new created genome mapping list. For example: Osp1g00110 AU075862 PFG_3A-08906.R or query1 chloroplast 0.0 W/1234-2345 search2 chr01 0.0 C/1234-2345,3456-4567 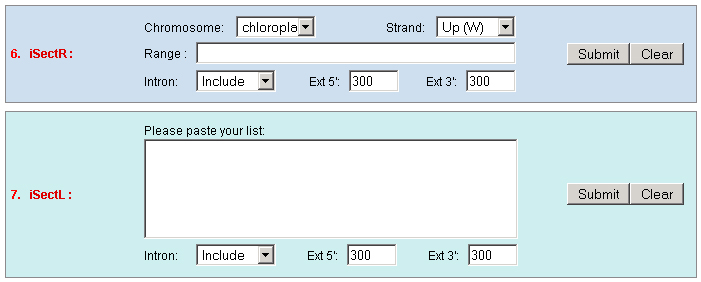 8. PrimerL: iSect primer (L) tool to design primers for a list of data names existed or a list of genome mapping data. It includes actually two steps: first to retrieve the genome sequences by Primer3.iSect tool and then design primers by the Primer3. a. Protocol to design primer pair for insertion. 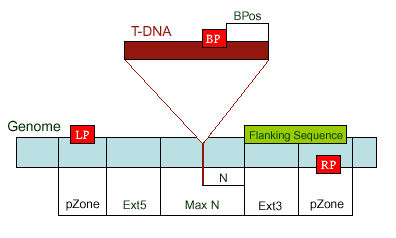 b. Protocol to design primer pair for target region or deletion. 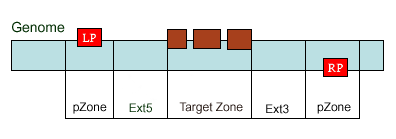 For example: Osp1g00110 AU075862 PFG_3A-08906.R or query1 chloroplast 0.0 W/1234-2345 search2 chr01 0.0 C/1234-2345,3456-4567 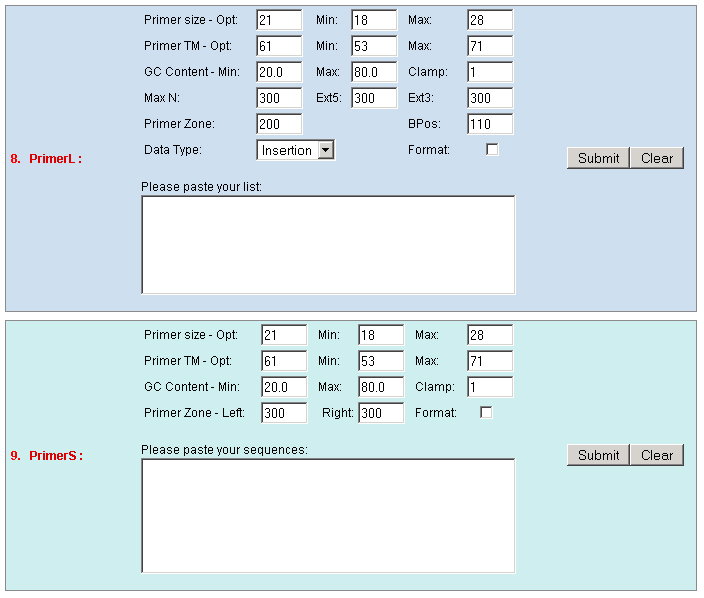 9. PrimerS: iSect primer (S) tool to design primers from user input sequences. For example: >YourInput.fa CTCGCTGGCCTGTTCAGGTGAATCTTAGTATCGTTTTGTGTCCATGTTGCCTCTACTTCG CCTGCTTCTGGTGACTTGAATTCTCTCTTTGTTTTGGGCAGCTTCGGGAACCTTCCACAT CCGAAGATGCTGAATTCGCTTCTATTTTGCGGACTCTCAAGGACAGTTTTGATAACGCAT TCACTGCTATGATTTCCTTCCAGACGAAGAATTCCGAGCGCATATTCAGCACAGGTATAA 10. GeneAtlas: Gene express atlas query tool by probe name or gene name. For example: Query: You can select an expression Data type, which is usually different in array type or platform
like GPL2025 [Data Type], and then input probe set or gene name, like AFFX-BioB-3_at or
Os01g01100 in the Query field. RMOS [Data Type] 001-200-E04 or Os07g04850 Filter: You can input words to select the experiments which contain your keywords. For example:
GSE7951 GSE7952 or root Seedling ABA (allow maximum 5 words). PageSize: Number of the display samples in a page.  |